pre-mRNA splicing
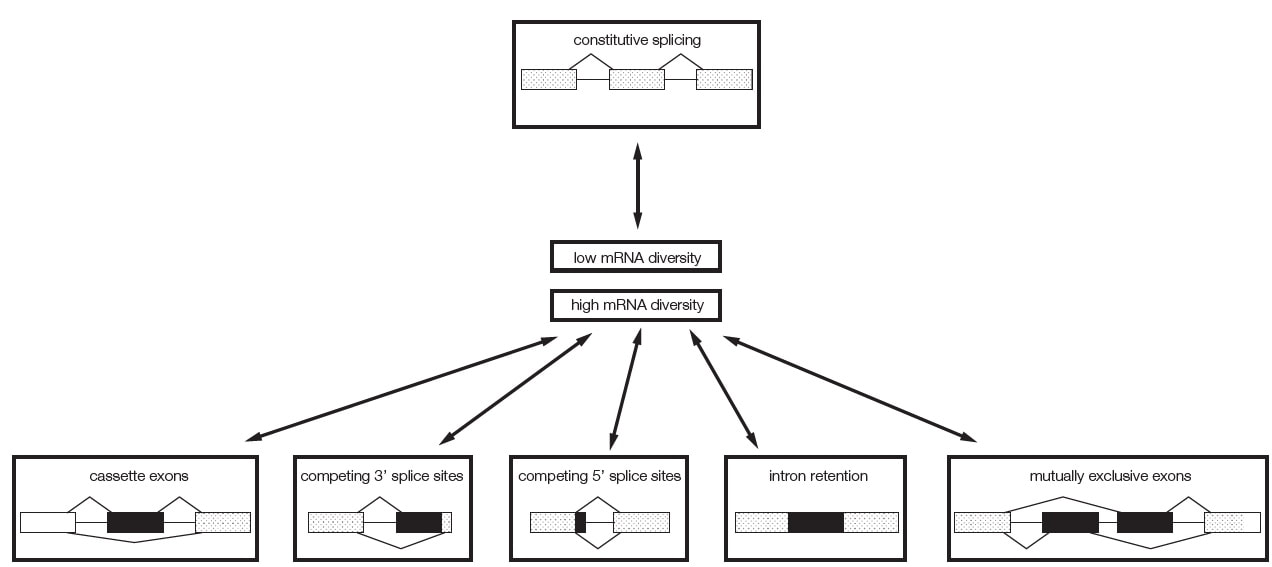
pre-mRNA splicing is a maturation event in which parts of the pre-mRNA sequences are joined and exported into the cytosol as exons and the intervening sequences are removed as introns. Alternative pre-mRNA splicing is the alternate inclusion of exons or introns from the primary transcript (pre-mRNA) into mature mRNA. The majority of human genes are alternatively spliced, i.e. contain sequences that can be used either as an exon or as an intron. Alternative splicing creates different mRNA isoforms, from a single gene. Mostly, but not always, these mRNA isoforms encode distinct protein products.
Alternative splicing increases the number of RNA (and protein isoforms) made from the genome.
Alternative splicing increases the number of RNA (and protein isoforms) made from the genome.
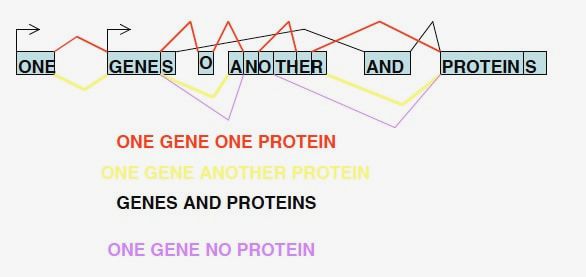
This example shows how a combination of different splicing patterns can create many different and opposing messages from a single gene.
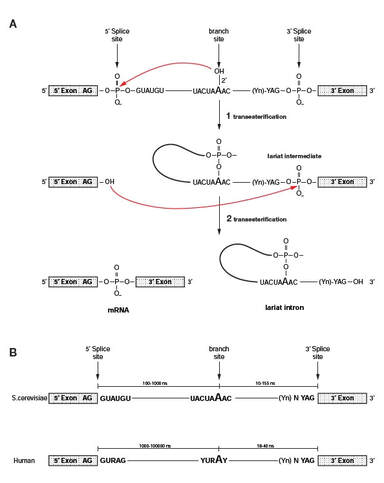
Chemistry of splicing
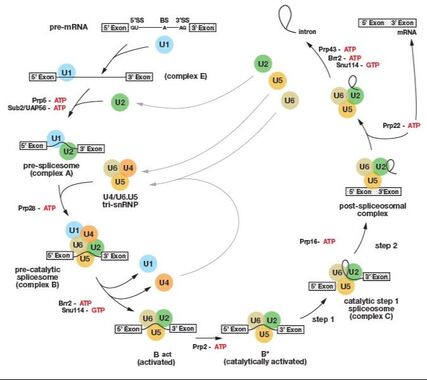
Splicing occurs in two consecutive transesterification reactions, catalyzed by the spliceosome. The backbone of the spliceosome are protein-RNA particles (U1, U2, U4/5/6) that perform the reaction.
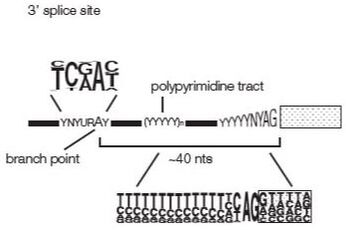
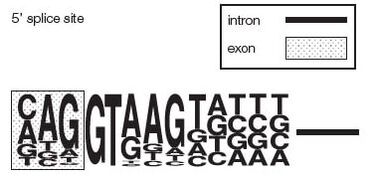
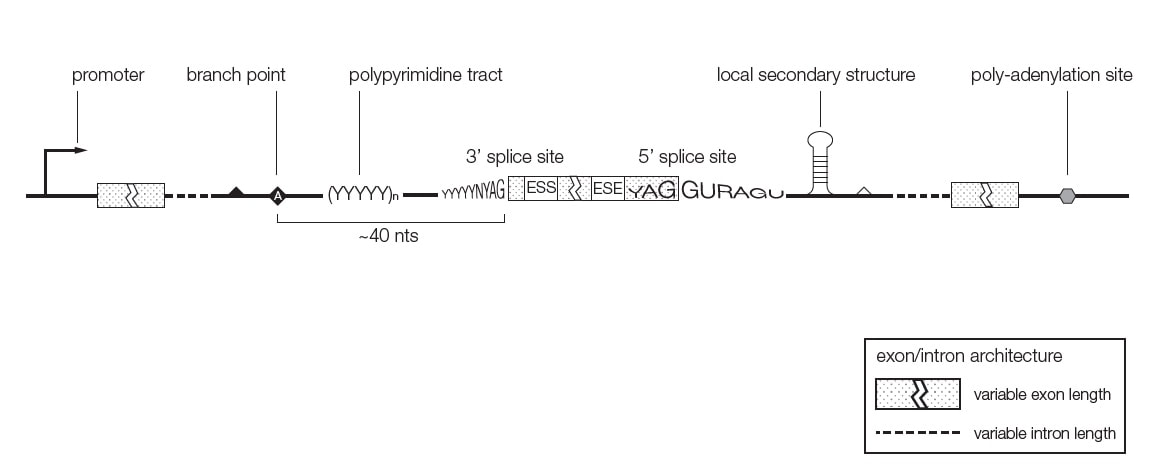
cis elements and trans-acting factors governing splice site selection
Exons surrounded by 5' and 3' splice site that follow a consensus sequence. Almost 100% conserved is the presence of AG and GT flanking internal exons, i.e. ag -EXON-gt.
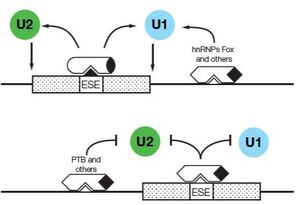
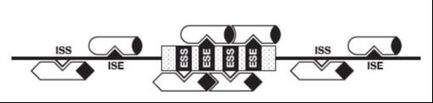
In addition to the splice sites, RNA elements in exons and introns contribute to exon recognition. These elements can act as silencers or enhancers, and can be located in introns and exons. In general, proteins bound to splicing enhancers attract components of the splicesome that contain small RNAs that bind to pre-mRNA splice sites. Proteins bound to silencers antagonize these interactions. The RNA elements are typically short 4-6 nt and bind to proteins. The proteins interact with each other, forming transient complexes on the pre-mRNA that help exon recognition.
As a result of multiple, weak protein-RNA, RNA-RNA and protein-protein interactions (alternative) exon recognition is influenced by multiple factors: polymerase speed, adherence of RNA elements to consensus sequences, secondary structures and concentration/phosphorylation state of regulator factors.