Overview
The long-term aim of our research is to understand the regulation and function of alternative pre-mRNA processing, mainly alternative splicing, but also regulation of RNA stability, and the formation of circular RNAs. We apply the knowledge gained in designing therapies against diseases caused by errors in RNA processing, such as Prader-Willi and Alzheimer’s disease.
1. Role of circular RNAs in Alzheimer’s disease
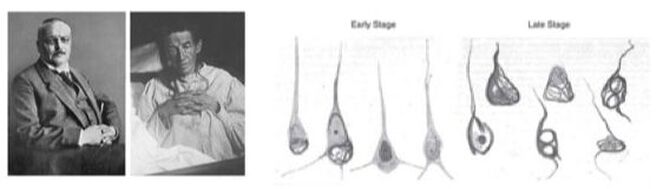
The microtubule associated protein tau plays a central role in neurodegenerative diseases, most prominent in Alzheimer’s disease. In the diseased state, the Tau protein can mis-fold into protein aggregates (paired helical filaments and neurofibrillary tangles), which likely cause neuronal death and are characteristic histological findings in Alzheimer’s disease (Figure 1).
Figure 1: Left: Alois Alzheimer and his famous patient Auguste Deter. Right: Alzheimer’s drawings of neurons showing tau deposits. Description of Auguste Deter in Alzheimer A. Über eine eigenartige Erkrankung der Hirnrinde. Allgemeine Zeitschrift fur Psychiatrie und Psychisch-gerichtliche Medizin. 1907 Jan;64:146-8, translated in PMID: 8713166 and Alzheimer A. Über eigenartige Krankheitsfälle des späten Alters. Z Gesamte Neurol. Psychiatr. 1911(4):356–85.
The tau protein is evolutionary well-conserved and almost identical between human and mouse, making it difficult to understand why humans, but not mice, are affected by Alzheimer’s. However, there are striking difference on the genomic level. The human tau gene contains short repetitive DNA elements (Alu elements) in its introns, i.e. parts of the gene that are cut out and removed before proteins are made. These Alu elements originated in primates and increased in number during evolution. From all species, humans have the highest number of Alu elements in the tau locus. Due to their structure, Alu elements bind to each other.
The presence of 83 Alu elements in the tau pre-mRNA gives it a unique, human-specific structure, as Alu elements bind to each other. As a result of this structure, the human tau gene generates human-specific circular RNAs that we discovered.
These circular tau RNAs encode proteins and we investigate whether these proteins are translated and form ‘seeds’ for tau protein aggregates (Figure 2).
We are thus testing the hypothesis that protein encoded by human-specific circular tau RNAs, not the proteins encoded by the linear RNAs are the basis of tau deposits resulting in Alzheimer’s disease. We are thus determining molecular events that promote circular RNA formation and their translation into proteins.
The presence of 83 Alu elements in the tau pre-mRNA gives it a unique, human-specific structure, as Alu elements bind to each other. As a result of this structure, the human tau gene generates human-specific circular RNAs that we discovered.
These circular tau RNAs encode proteins and we investigate whether these proteins are translated and form ‘seeds’ for tau protein aggregates (Figure 2).
We are thus testing the hypothesis that protein encoded by human-specific circular tau RNAs, not the proteins encoded by the linear RNAs are the basis of tau deposits resulting in Alzheimer’s disease. We are thus determining molecular events that promote circular RNA formation and their translation into proteins.
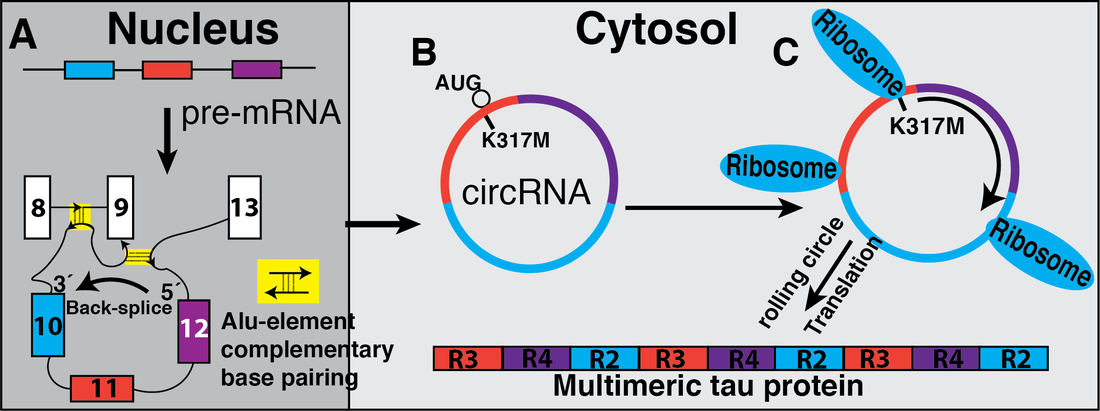
Figure 2: A. Tau pre-mRNA contains Alu-elements that help in the formation of circular structures and promote backsplicing between exon 10 (blue) and 12 (purple). B. The circular RNAs are exported to the cytosol. C. The circRNA 1210 with FTDP-17, K317M mutation creates start codon allowing translation forming multimeric proteins that can potentially contribute to neurofibrillary tau tangles and the disease pathology. CircRNA 127 has endogenous in frame start codon with no stop codon and is translated under the same circumstances.
Key reference:
Welden JR, van Doorn J, Nelson PT, Stamm S. The human MAPT locus generates circular RNAs. Biochim Biophys Acta Mol Basis Dis. 2018:2753-60.PubMed PMID: 29729314.
Welden JR, van Doorn J, Nelson PT, Stamm S. The human MAPT locus generates circular RNAs. Biochim Biophys Acta Mol Basis Dis. 2018:2753-60.PubMed PMID: 29729314.
2. Role of non-coding RNAs (snoRNAs) in Prader-Willi syndrome
We study the molecular cause for Prader-Willi syndrome, the most common genetic cause for systemic obesity. The goal of the project is to a) understand the function of a new class of non-coding regulatory RNAs and b) develop therapies against hyperphagia and obesity.
Prader-Willi syndrome is caused by the loss of expression of the ‘Prader-Willi critical region’, located on chromosome 15. Recently, several patients with Prader-Willi like phenotypes have been described. These patients had microdeletions affecting two clusters of small nucleolar RNAs, SNORD115 and SNORD116, indicating that the loss of small nucleolar RNAs plays a central role in the etiology of the disease.
We are determining the molecular mechanisms that link the loss of SNORD115 and SNORD116 to the syndrome and obesity. Loss of SNORD115 could explain hyperphagia observed in PWS patients, since SNORD115 was shown to regulate alternative splicing of the serotonin receptor 2C causing generation of the most active form of the receptor. SNORD116 influences the expression of miRNAs hosted in the second exon of the serotonin receptor 2C and we currently investigating the influence of SNORD116 on gene expression.
SNORD115 and SNORD116 do not express canonical C/D box snoRNAs, but are likely processed into shorter RNAs, termed psnoRNAs that do not associate with the proteins typically found in C/D box snoRNAs. Canonical C/D box snoRNAs function in 2’O methylation of non-coding RNAs and form a defined ribonuclear complex with the proteins fibrillarin, NOP56/58 and 15.5kD (NHP2L1), (see review about processed snoRNAs).
Thus SNORD116 and SNORD115 generate a new class of regulatory RNAs that regulate RNA processing. We are currently determining their target genes and molecular mechanism. We identified drugs that similar to SNORD115 promote formation of the most active serotonin receptor 2C isoform, which are tested in mice.
Prader-Willi syndrome is caused by the loss of expression of the ‘Prader-Willi critical region’, located on chromosome 15. Recently, several patients with Prader-Willi like phenotypes have been described. These patients had microdeletions affecting two clusters of small nucleolar RNAs, SNORD115 and SNORD116, indicating that the loss of small nucleolar RNAs plays a central role in the etiology of the disease.
We are determining the molecular mechanisms that link the loss of SNORD115 and SNORD116 to the syndrome and obesity. Loss of SNORD115 could explain hyperphagia observed in PWS patients, since SNORD115 was shown to regulate alternative splicing of the serotonin receptor 2C causing generation of the most active form of the receptor. SNORD116 influences the expression of miRNAs hosted in the second exon of the serotonin receptor 2C and we currently investigating the influence of SNORD116 on gene expression.
SNORD115 and SNORD116 do not express canonical C/D box snoRNAs, but are likely processed into shorter RNAs, termed psnoRNAs that do not associate with the proteins typically found in C/D box snoRNAs. Canonical C/D box snoRNAs function in 2’O methylation of non-coding RNAs and form a defined ribonuclear complex with the proteins fibrillarin, NOP56/58 and 15.5kD (NHP2L1), (see review about processed snoRNAs).
Thus SNORD116 and SNORD115 generate a new class of regulatory RNAs that regulate RNA processing. We are currently determining their target genes and molecular mechanism. We identified drugs that similar to SNORD115 promote formation of the most active serotonin receptor 2C isoform, which are tested in mice.

Figure 3: The image depicts Doña Eugenia Martinez Vellejo, a likely Prader Willi Syndrome (PWS) subject, by the Spanish baroque painter Juan Carreño de Miranda (1614–1685). Doña Eugenia was approximately six years of age at the time of the painting (c. 1680).1 The maternally imprinted region on chromosome 15 that is involved in PWS is shown. Genes expressed from the paternal chromosome are in yellow, gene expressed from the maternal chromosome are in blue, genes expressed from both chromosomes are in gray. PWS is caused by the loss of gene expression from the paternal chromosome. BP1, BP2 and BP3 are the breakpoint cluster regions. IC is the imprinting center.
Expression units hosting small nuclear RNAs (snoRNAs) are shown as thin vertical lines. In humans, there are at least 24 and 47 copies of the SNORD 116 (HBII-85) and SNORD115 (HBII-52) units, respectively. One of these units is enlarged below the gene structure and shows the homing exons in yellow, and the snoRNA in black. These snoRNAs give rise to smaller RNAs (psnoRNAs for processed snoRNA) that regulate alternative splicing (see Kishore et al., 2010).
1Pozzilli, P. and Khazrai, Y.M. (2005) "La Monstrua Vestida", a case of Prader-Willi syndrome. J Endocrinol Invest, 28, 199.
Expression units hosting small nuclear RNAs (snoRNAs) are shown as thin vertical lines. In humans, there are at least 24 and 47 copies of the SNORD 116 (HBII-85) and SNORD115 (HBII-52) units, respectively. One of these units is enlarged below the gene structure and shows the homing exons in yellow, and the snoRNA in black. These snoRNAs give rise to smaller RNAs (psnoRNAs for processed snoRNA) that regulate alternative splicing (see Kishore et al., 2010).
1Pozzilli, P. and Khazrai, Y.M. (2005) "La Monstrua Vestida", a case of Prader-Willi syndrome. J Endocrinol Invest, 28, 199.
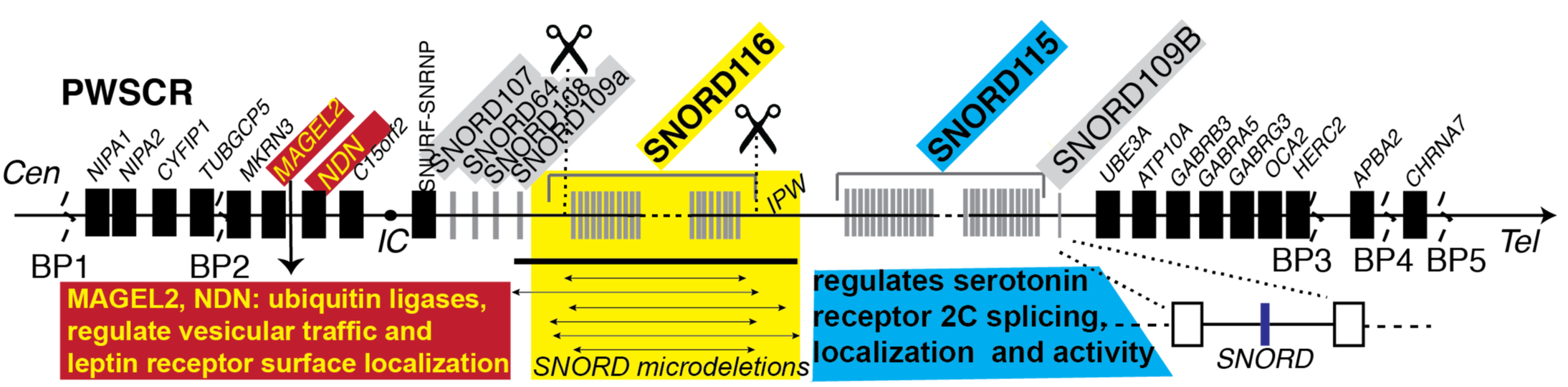
Figure 4: SNORDs contribute to Prader-Willi syndrome
The Prader-Willi critical region is defined by breakpoints (BP, BP1-BP5). A. MAGEL2 loss or mutations lead to obesity less severe than in PWS. B. Deletion of the six “SNORD microdeletions” encompassing the SNORD116 cluster result in a PWS-like phenotype. Scissors indicated location of our CRISPR clones. C. SNORD115 loss leads to abarrant serotonin receptor 2C splicing decreasing receptor surface abundance resulting in obesity. D. Each SNORD expression unit is composed of two non-coding exons (boxes) flanking the intron containing the SNORD (blue line).
The Prader-Willi critical region is defined by breakpoints (BP, BP1-BP5). A. MAGEL2 loss or mutations lead to obesity less severe than in PWS. B. Deletion of the six “SNORD microdeletions” encompassing the SNORD116 cluster result in a PWS-like phenotype. Scissors indicated location of our CRISPR clones. C. SNORD115 loss leads to abarrant serotonin receptor 2C splicing decreasing receptor surface abundance resulting in obesity. D. Each SNORD expression unit is composed of two non-coding exons (boxes) flanking the intron containing the SNORD (blue line).
Key references
Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006; 311(5758):230-2.PubMed PMID: 16357227.
Falaleeva M, Pages A, Matuszek Z, Hidmi S, Agranat-Tamir L, Korotkov K, Nevo Y, Eyras E, Sperling R, Stamm S. Dual function of C/D box snoRNAs in rRNA modification and alternative pre-mRNA splicing Proc Natl Acad Sci U S A. 2016;113(12):E1625-34.PubMed PMID: 26957605.
Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006; 311(5758):230-2.PubMed PMID: 16357227.
Falaleeva M, Pages A, Matuszek Z, Hidmi S, Agranat-Tamir L, Korotkov K, Nevo Y, Eyras E, Sperling R, Stamm S. Dual function of C/D box snoRNAs in rRNA modification and alternative pre-mRNA splicing Proc Natl Acad Sci U S A. 2016;113(12):E1625-34.PubMed PMID: 26957605.
3. Changing alternative splicing of the serotonin receptor 2C to combat spasticity and obesity
The major target of SNORD115 missing in PWS is the serotonin receptor 2C (5HT2C). 5HT2C regulates food uptake and muscle tone. Its activity is regulated by alternative pre-mRNA splicing as this mechanism generates a truncated receptor protein isoform that sequesters the full-length receptor in intracellular membranes. We developed an oligonucleotide that promotes exon inclusion, which increases the ratio of the full-length to truncated receptor protein. This oligo reduces food intake after injection in mice (Figure 5).
Another dysregulation of the 5HT2C isoforms is observed after spinal cord injury, where a change in 5HT2C isoforms causes spasms. We using oligonucleotides that change the splicing pattern and are testing them in rat models of spinal cord injury.
Another dysregulation of the 5HT2C isoforms is observed after spinal cord injury, where a change in 5HT2C isoforms causes spasms. We using oligonucleotides that change the splicing pattern and are testing them in rat models of spinal cord injury.
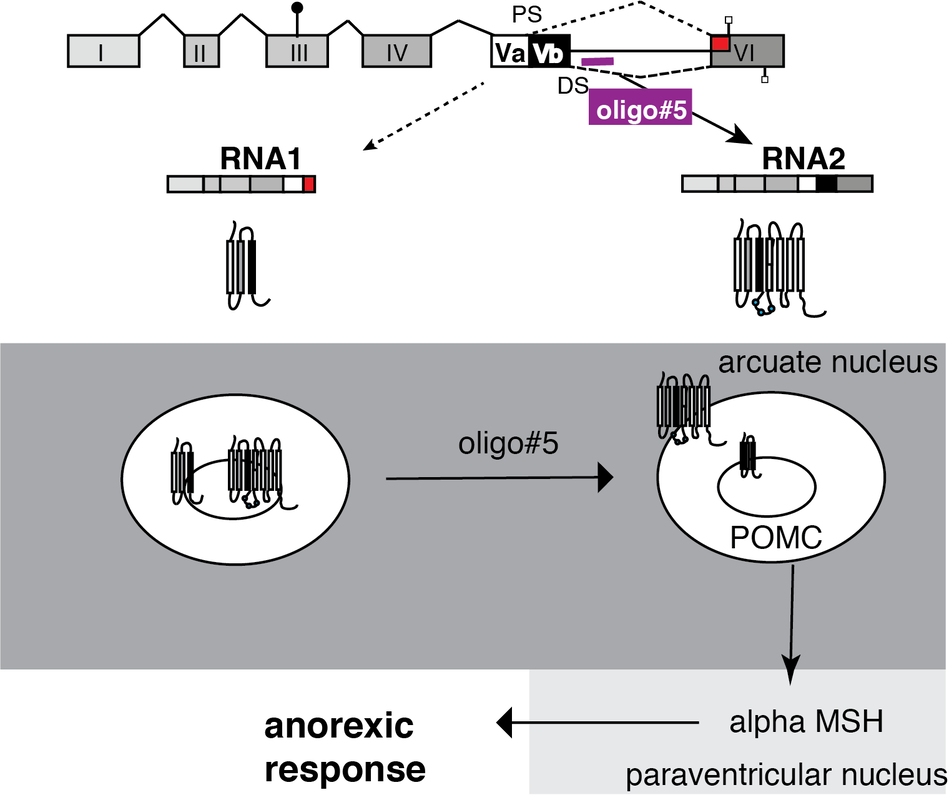
Figure 5: The activity of the serotonin receptor 2C is regulated by alternative splice variants and can be influenced by oligonucleotides.
The 5HT2C receptor generates two major RNA isoforms generating a truncated receptor and a full-length receptor. The truncated isoform is retained in intracellular membranes. Due to heterodimerization, it sequesters the full-length receptor inside the cell. The loss of the truncated receptor isoform caused by oligo#5 releases this sequestration and causes an accumulation of the receptor on the cell surface, which results in an anorexic response.
The 5HT2C receptor generates two major RNA isoforms generating a truncated receptor and a full-length receptor. The truncated isoform is retained in intracellular membranes. Due to heterodimerization, it sequesters the full-length receptor inside the cell. The loss of the truncated receptor isoform caused by oligo#5 releases this sequestration and causes an accumulation of the receptor on the cell surface, which results in an anorexic response.
Key reference
Zhang Z, Shen M, Gresch P, Ghamari-Langroudi M, Rabchevsky AG, Emeson RB, Stamm S. Oligonucleotide-induced alternative splicing of serotonin 2C receptor reduces food intake. EMBO molecular medicine. 2016;8(8):878-94.PubMed PMID: 27406820.
Zhang Z, Shen M, Gresch P, Ghamari-Langroudi M, Rabchevsky AG, Emeson RB, Stamm S. Oligonucleotide-induced alternative splicing of serotonin 2C receptor reduces food intake. EMBO molecular medicine. 2016;8(8):878-94.PubMed PMID: 27406820.
4. Identifying siRNAs that target circular RNAs